All Services
Sequencing Services
De novo whole genome sequencing (WGS) is a method to reconstruct an entire genome of an organism without prior knowledge of its sequence, through the assembly of short and long fragments of DNA, generated using next-generation sequencing technologies.
Whole genome re-sequencing (WGR) involves sequencing an individual’s entire genome and comparing it to a reference genome to identify genetic differences such as SNPs, indels, translocations, inversions, and CNVs.
Whole exome sequencing (WES) is a cost-effective method that analyzes only the protein-coding regions of the genome. While these regions comprise only 2% of the genome, they are responsible for coding for proteins that make up to 85% of known disease-causing variants.
Metagenomics examines the genetic content of microbiota in a given environment using a shotgun sequencing approach on next-generation sequencing platforms. Metagenomics provides comprehensive pictures of microbial diversity, genetic relationships, and functional activities within and between microbial communities.
Double digest genotyping by sequencing (ddGBS) is a cost-effective method that uses two restriction enzymes with NGS to discover and genotype SNPs without prior knowledge of the genomic sequence. It is the preferred strategy for high-throughput SNP discovery and genotyping in non-model organisms.
Messenger RNA sequencing (mRNAseq) is a widely used tool for studying gene transcripts, quantifying gene expression changes, and providing an unbiased view of the transcriptional landscape without a need of a reference genome. Applicable to eukaryotes and prokaryotes.
Long non-coding RNA sequencing (lncRNAseq) is a next generation sequencing method used to detect expression of lncRNAs and mRNAs, identify novel RNA and splice junctions, and provides valuable insights on the regulatory mechanisms of non-coding RNAs.
Small non-coding RNA sequencing (sncRNAseq) is a technique to detect and quantify small non-coding RNAs (<200nt), including miRNAs, siRNAs, snRNAs, among others. Some of their functions include gene silencing, post-transcriptional gene regulation, splicing and RNA processing.
Whole transcriptome sequencing (WTS) is a combination of lncRNAseq and sncRNAseq, used to identify and quantify mRNA, long and small non-coding RNA. It provides the most comprehensive view of the global transcripts profile.
Meta-transcriptomics uses RNAseq to examine gene expression in microorganisms in a given environment. Meta-transcriptomics can provide insights to biomarkers, taxonomic composition, and metabolic processes and functional profiles.
Whole genome bisulfite sequencing (WGBS) is a method for detecting methylated cytosines in genomic DNA, providing single-base resolution of methylation status across the entire genome. DNA methylation of cytosine is important in gene expression and chromatin remodeling, and abnormal patterns are associated with diseases.
Chromatin immunoprecipitation combined with next generation sequencing (ChIPseq) is a method for identifying genome-wide DNA binding sites for transcription factors and other proteins. ChIP-Seq is used to map DNA-binding proteins and histone modifications, at base-pair resolution, on a genome-wide scale.
Amplicon sequencing is a highly targeted technique using primers to generate PCR products (amplicons) followed by next generation sequencing, to analyze genetic variation in specific genomic regions. Amplicon sequencing is used for genome targeting and detecting variations, copy number variations, gene fusions, InDels and SNPs.
Metabarcoding is a form of amplicon sequencing that amplifies unique DNA barcodes using PCR followed by high-throughput sequencing to identify multiple taxa and detect novel species in a given environment. DNA barcodes such as 16S, 18S and ITS are routinely used in metabarcoding.
Multiplex PCR-based targeted amplicon sequencing (MTAseq) use multiplex PCR to amplify tens to thousands of target regions in a single PCR reaction, followed by next generation sequencing. This approach allows for the simultaneous analysis of large numbers of target genes.
Sample Management
Freezer Pro
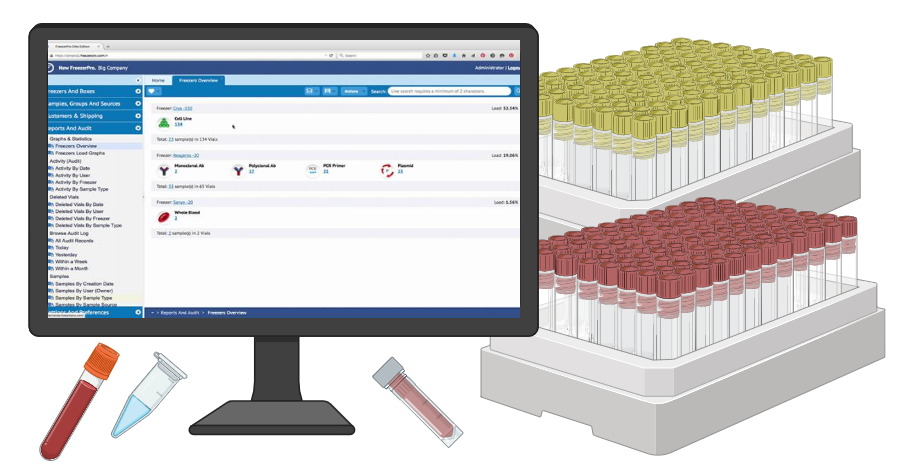
FreezerPro is a sample management software that streamlines sample tracking. With its advanced features, it maximizes precision and efficiency for research institutes and pharmaceutical labs worldwide. FreezerPro boasts an intuitive interface that offers a rapid and dependable method of finding any sample's location. Over 1,000 organizations have subscribed to and trust FreezerPro for their daily sample management operations. Become a part of the FreezerPro community and alleviate the tedious process of searching for samples.